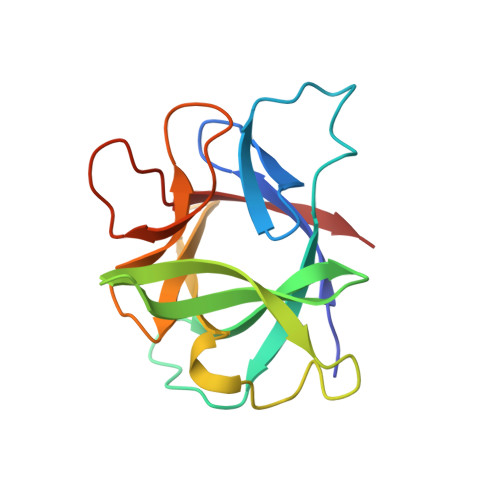
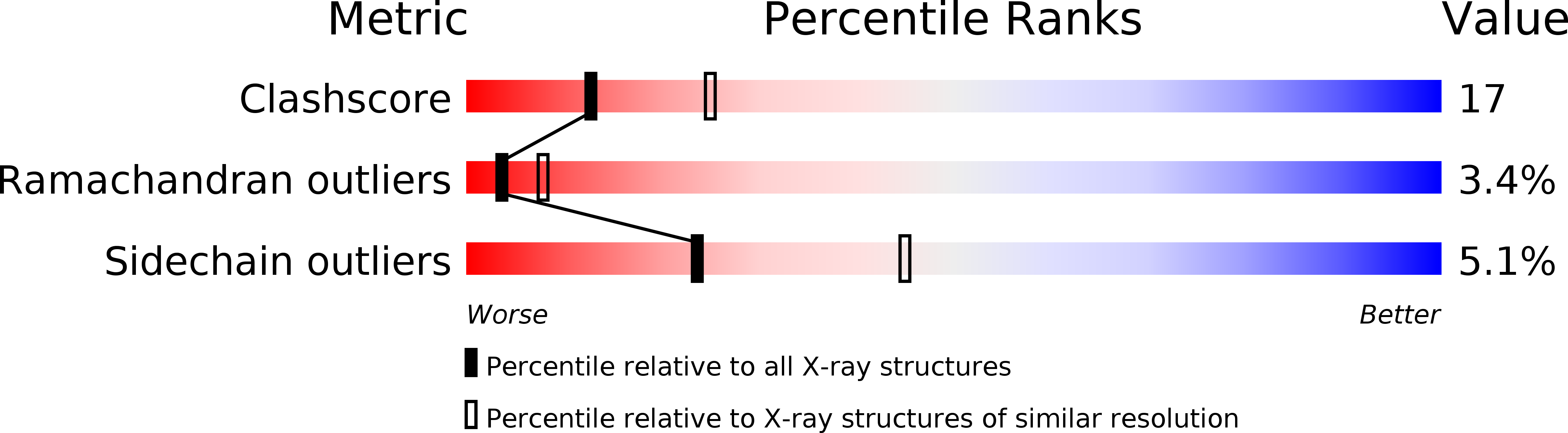The structure of murine interleukin-1 beta at 2.8 A resolution.
van Oostrum, J., Priestle, J.P., Grutter, M.G., Schmitz, A.(1991) J Struct Biol 107: 189-195
- PubMed: 1807351
- DOI: https://doi.org/10.1016/1047-8477(91)90021-n
- Primary Citation of Related Structures:
2MIB - PubMed Abstract:
The three-dimensional structure of recombinant murine interleukin-1 beta has been solved by X-ray crystallographic techniques to 2.8 A resolution and refined to a crystallographic R factor of 0.192. Although murine interleukin-1 beta crystallizes in the same space group as human interleukin-1 beta with almost identical unit cell dimensions, the packing of the molecules is quite different. The murine interleukin-1 beta structure was solved by molecular replacement using the refined structure of human interleukin-1 beta as trial structure, and found to be related to the human structure by a nearly perfect twofold rotation about the crystallographic y-axis and a 14 degrees rotation about the z-axis, with no translation. The folding of murine interleukin-1 beta is similar to that found for the human variant, consisting of 12 beta strands wrapped around a core of hydrophobic side chains in a tetrahedron-like fashion. Significant differences with respect to the human structure are seen at the N terminus and in 4 of the 11 loops connecting the 12 beta strands.
Organizational Affiliation:
Department of Biotechnology, Ciba-Geigy Ltd., Basel, Switzerland.